Ongoing Projects
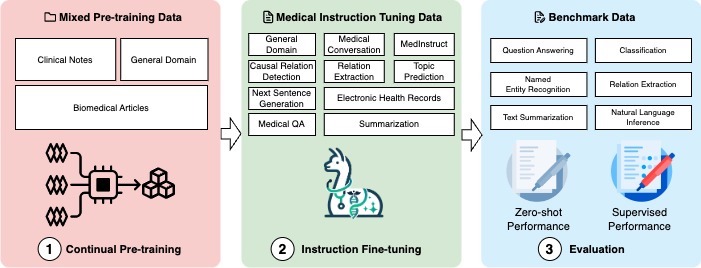
Foundation Large Language Models
Our lab is a pioneer in harnessing advanced Language Models (LLMs), including but not limited to LLaMA and GPT, to extract valuable insights from diverse medical texts. From electronic health records to scientific literature, we employ these models to decode complex medical narratives and uncover hidden knowledge.
Unlike other LLMs, our models are specifically tailored to the biomedical domain with comprehensive domain-specific training data. This allows us to develop models that are more accurate and efficient in understanding and generating biomedical texts. Read more
DataMed
DataMed is a biomedical data search engine. Its goal is to discover data sets across data repositories or data aggregators. In the future it will allow searching outside these boundaries. DataMed supports the NIH-endorsed FAIR principles of Findability, Accessibility, Interoperability and Reusability of datasets with current functionality assisting in finding datasets and providing access information about them.


Biomedical Knowledge Explorer
In the rapidly evolving field of biomedicine, researchers are inundated with an ever-growing corpus of publications, including research scientific articles, books, technical reports, and working papers. This massive biomedical knowledge poses a significant challenge in terms of efficient data navigation and exploration. While several search engines have been developed to improve the search and browsing of massive literature, gaps in efficient knowledge exploration and global understanding persist.
To address this gap, we propose BIKE (Biomedical Knowledge Explorer), a visual analytics tool leveraging a large language model approach to explore the semantic embedding space of massive publications. By combining advanced language processing techniques with intuitive visualizations, BIKE offers researchers a powerful means to navigate and comprehend the complex landscape of biomedical literature.
NLP for Real World Studies
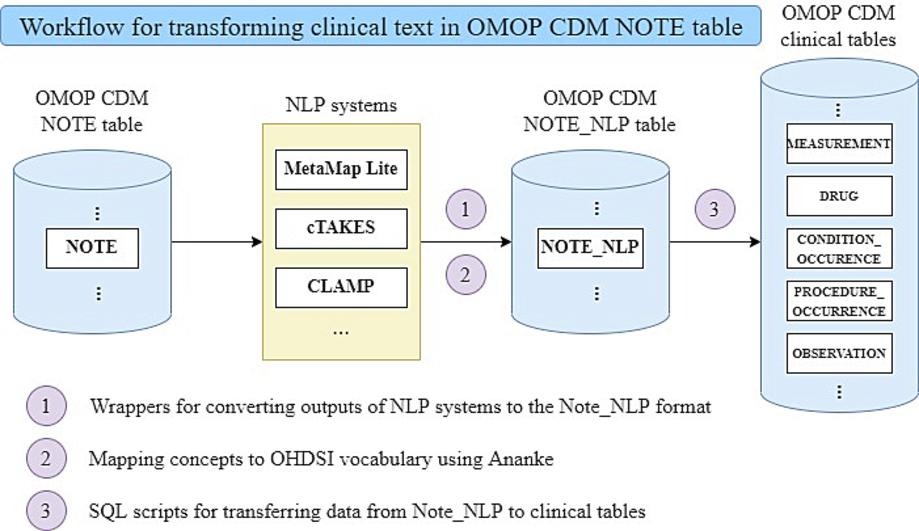
Clinical documentation in electronic health records contains crucial narratives and details about patients and their care. Natural language processing (NLP) can unlock the information conveyed in clinical notes and reports, and thus plays a critical role in real-world studies. The NLP Working Group at the Observational Health Data Sciences and Informatics (OHDSI) consortium was established to develop methods and tools to promote the use of textual data and NLP in real-world observational studies. In this paper, we describe a framework for representing and utilizing textual data in real-world evidence generation, including representations of information from clinical text in the Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM), the workflow and tools that were developed to extract, transform and load (ETL) data from clinical notes into tables in OMOP CDM, as well as current applications and specific use cases of the proposed OHDSI NLP solution at large consortia and individual institutions with English textual data. Challenges faced and lessons learned during the process are also discussed to provide valuable insights for researchers who are planning to implement NLP solutions in real-world studies. Read more
Grants
Current
-
U24LM013755 (12/21/2020 - 11/30/2024): RADx-Rad Discoveries & Data: Consortium Coordination Center Program Organization
-
R56AG069880 (12/21/2020 - 11/30/2024): Advancing Drug Repositioning for Alzheimer’s Disease using Real-world Data
-
RF1AG072799 (5/1/2021 - 4/30/2024): Facilitate Observational Studies of Alzheimer's Disease and Alzheimer's Disease-Related Dementias Using Ontology and Natural Language Processing
-
R01LM013519 (9/1/2021 - 5/31/2025): PheBC: bias correction methods for EHR derived phenotype
-
R01AG073435 (9/15/2021 - 5/31/2026): TRiPOD: Toward Reusable Phenotypes in Observational Data for AD/ADRD - managing definitions and correcting bias
-
R01AG078154 (9/1/2022 - 5/31/2027): Detecting synergistic effects of pharmacological and non-pharmacological interventions for AD/ADRD
-
U24MH130988 (9/1/2022 - 6/30/2027): Engagement and outreach to achieve a FAIR data ecosystem for the BICAN
-
R01AG080429 (2/15/2023 - 1/31/2028): Leveraging Longitudinal Data and Informatics Technology to Understand the Role of Bilingualism in Cognitive Resilience, Aging and Dementia
Past
-
U24CA194215 (09/01/2016 - 08/31/2023): Advancing Cancer Pharmacoepidemiology Research through EHRs and Informatics
-
69357270 (10/01/2015 - 09/30/2018): pSCANNER (patient-centered SCAlable National Network for Effectiveness Research)
-
U2COD023196 (07/01/2016 - 06/30/2021): Partnership in Learning around Engagement, Data, Genomics, and Environment
-
U24CA194215 (09/01/2016 - 08/31/2021): Advancing Cancer Pharmacoepidemiology Research through EHRs and Informatics
-
R01LM011829 (09/01/2014 - 08/31/2018): Patient Medical History Representation, Extraction, and Inference from EHR Data
-
R01HS022895 (09/30/2014 - 09/29/2019): Learning from patient safety events: A case based toolkit
-
R01LM010681 (05/31/2010 - 09/28/2018): Interactive machine learning methods for clinical natural language processing
-
R01GM103859 (09/18/2014 – 5/31/2018): Informatics Tools for Pharmacogenomic Discovery using Practice-based Data
-
R01GM102282 (04/01/2013 - 03/31/2017): Natural Language Processing for Clinical and Translational Research
-
U24AI117966 (09/29/2014 - 08/31/2017): BioCADDIE: Biomedical and healthCAre Data Discovery and Indexing Ecosystem
-
U01CA180964 (09/01/13 - 08/31/16): Informatics to enable routine personalized cancer therapy.
-
R01LM011563 (09/01/13 - 08/31/16): Using Biomedical Knowledge to Identify Plausible Signals for Pharmacovigilance